New technology maps where and how cells read their genome
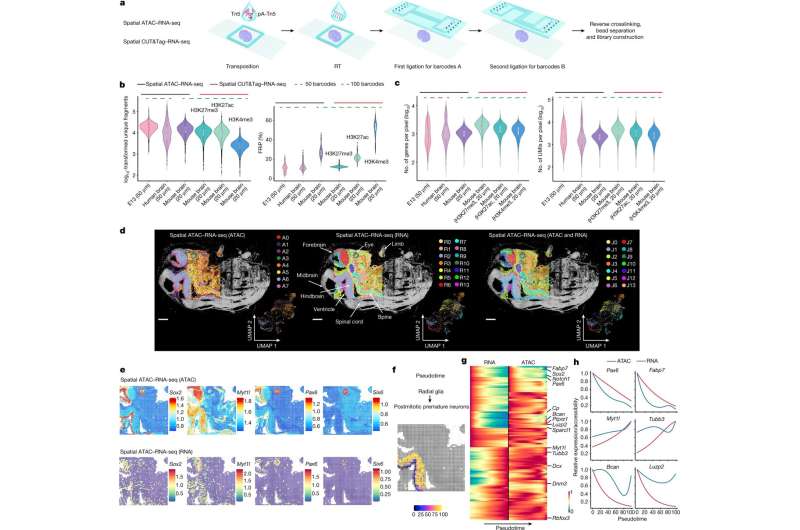
A new study revealed in Character reviews that a engineering identified as spatial omics can be utilised to map concurrently how genes are switched on and off and how they are expressed in distinctive spots of tissues and organs. This enhanced engineering, designed by scientists at Yale College and Karolinska Institutet, could drop gentle on the improvement of tissues, as effectively as on certain illnesses and how to address them.
Practically all cells in the system have the exact established of genes and can in theory turn into any kind of cell. What distinguishes the cells is how the genes in our DNA are made use of. In new yrs, spatial omics have supplied us a deeper being familiar with of how cells read through the genome in precise spots in tissues. Now, researchers have even more progressed this technology to enhance awareness of how tissues produce and how various illnesses come up.
A crucial component of the research is the researchers’ means to spatially map at the same time two vital parts of our genetic make-up, the epigenome and the transcriptome. The epigenome controls the switching mechanisms that switch genes on and off in particular person cells, whereas the transcriptome is the final result of people gene expressions and what makes just about every mobile exclusive.
Can detect input and output concurrently
The epigenome can be thought of as a fuse box. You can flip the switches, but if you can’t see irrespective of whether the lights are turning on, your data is limited. By spatially mapping each the epigenome and transcriptome, the researchers designed a technology in which both of those the input (switching on or off a gene) and the output (gene expression) can be detected in the very same tissue part. This new know-how delivers unparalleled insights into gene regulation in specific destinations in a tissue.
For this study, the researchers tailored and combined their earlier developed strategies to map the epigenome and the transcriptome, and used these tactics to mouse brains and human mind tissue.
“Now that we can merge the two, we can see both equally the mechanisms of how the genes are switched on and off, as nicely as the result,” suggests Gonçalo Castelo-Branco, professor at the Department of Health care Biochemistry and Biophysics, Karolinska Institutet, and one particular of the corresponding authors. “This has led us to some surprising observations, which offers us further more insights into how these processes are controlled in unique tissue places and contribute to distinctive cell fates.”
Advancing the area of personalised medicine
The get the job done could carry scientists closer to knowledge prospective genetic targets for drug remedy, and help progress the industry of individualized medication.
“In the long run with this technology, we will be able to really comprehend in each individual one client how those people cancer-promoting genes and tumor suppressor genes are staying controlled by the epigenetic mechanisms,” claims Rong Supporter, professor at Yale University and previous writer of the paper. “The whole epigenetic therapeutics field is just emerging, but I assume our technological innovation can possibly empower epigenetic drug discovery.”
Extra data:
Di Zhang et al, Spatial epigenome–transcriptome co-profiling of mammalian tissues, Mother nature (2023). DOI: 10.1038/s41586-023-05795-1
Citation:
New know-how maps exactly where and how cells read through their genome (2023, March 15)
retrieved 17 March 2023
from https://phys.org/news/2023-03-engineering-cells-genome.html
This doc is topic to copyright. Apart from any truthful dealing for the function of private examine or research, no
element may be reproduced without the prepared permission. The written content is offered for data functions only.
