MIT Turbocharges Python’s Notoriously Slow Compiler
Python has long been just one of—if not the—top programming languages in use. However when the significant-degree language’s simplified syntax would make it effortless to master and use, it can be slower in comparison to lessen-amount languages these types of as C or C++.
Researchers from MIT’s Personal computer Science and Artificial Intelligence Laboratory (CSAIL) hope to transform that by Codon, a Python-based mostly compiler that will allow buyers to publish Python code that operates as proficiently as a method in C or C++.
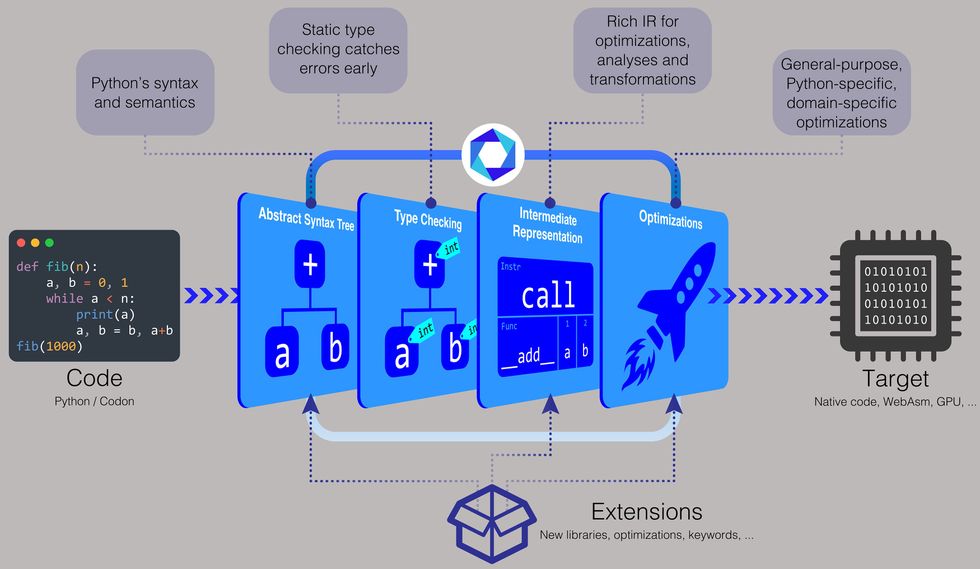
“Regular Python compiles to what is referred to as bytecode, and then that bytecode gets executed in a digital device, which is a great deal slower,” suggests Ariya Shajii, an MIT CSAIL graduate student and lead writer on a the latest paper about Codon presented in February at the 32nd ACM SIGPLAN Global Conference on Compiler Building. “With Codon, we’re executing indigenous compilation, so you’re managing the finish final result specifically on your CPU—there’s no intermediate digital machine or interpreter.”
The Python-based compiler arrives with pre-built binaries for Linux and macOS, and you can also build from source or create executables. “With Codon, you can just distribute the resource code like Python, or you can compile it to a binary,” Shajii claims. “If you want to distribute a binary, it will be the exact same as a language like C++, for illustration, where by you have a Linux binary or a Mac binary.”
To make Codon more quickly, the crew resolved to carry out sort examining in the course of compile time. Type examining requires assigning a facts type—such as an integer, string, character, or float, to name a few—to a benefit. For instance, the range 5 can be assigned as an integer, the letter “c” as a character, the word “hello” as a string, and the decimal variety 3.14 as a float.
“In normal Python, it leaves all of the types for runtime,” states Shajii. “With Codon, we do variety checking all through the compilation course of action, which allows us avoid all of that costly sort manipulation at runtime.”
MIT professor and CSAIL principal investigator Saman Amarasinghe, who’s also a coauthor on the Codon paper, provides that “if you have a dynamic language [like Python], each and every time you have some data, you require to preserve a large amount of more metadata all around it” to establish the style at runtime. Codon does absent with this metadata, so “the code is more quickly and knowledge is considerably more compact,” he suggests.
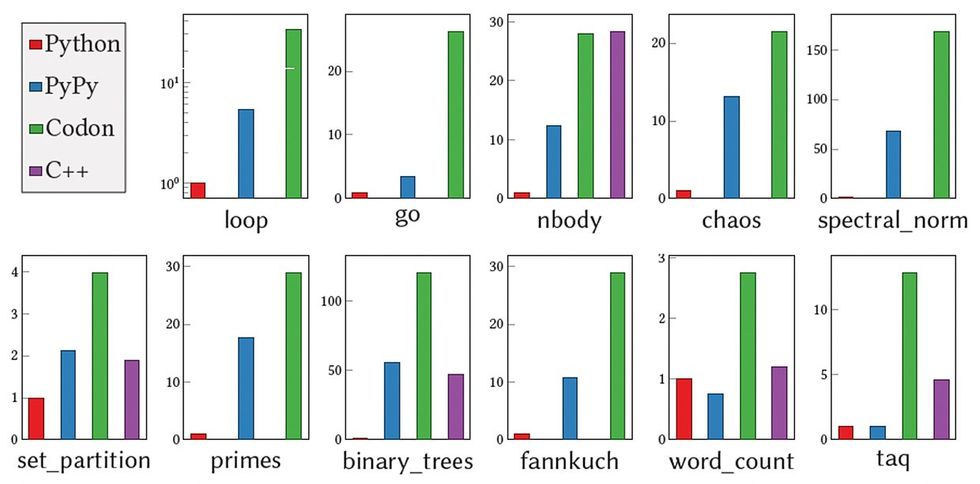
Devoid of any needless knowledge or variety examining through runtime, Codon results in zero overhead, in accordance to Shajii. And when it will come to overall performance, “Codon is usually on par with C++. As opposed to Python, what we typically see is 10 to 100x advancement,” he states.
But Codon’s solution will come with its trade-offs. “We do this static type checking, and we disallow some of the dynamic options of Python, like switching sorts at runtime dynamically,” states Shajii. “There are also some Python libraries we haven’t applied yet.”
Amarasinghe adds that “Python has been struggle-examined by quite a few folks, and Codon has not achieved anything like that but. It requires to run a great deal far more programs, get a great deal far more comments, and harden up much more. It will acquire some time to get to [Python’s] stage of hardening.”
Codon was to begin with made for use in genomics and bioinformatics. “Data sets are finding actually huge in these fields, and superior-amount languages like Python and R are way too gradual to take care of terabytes for each set of sequencing data,” says Shajii. “That was the gap we wished to fill—to give area professionals who are not automatically pc experts or programmers by coaching a way to tackle massive facts devoid of possessing to compose C or C++ code.”
Aside from genomics, Codon could also be applied to similar purposes that process enormous info sets, as nicely as spots this kind of as GPU programming and parallel programming, which the Python-based compiler supports. In reality, Codon is now staying utilized commercially in the bioinformatics, deep finding out, and quantitative finance sectors via the startup Exaloop, which Shajii established to shift Codon from an tutorial challenge to an industry application.
To allow Codon to function with these different domains, the workforce produced a plug-in method. “It’s like an extensible compiler,” Shajii states. “You can create a plug-in for genomics or a different domain, and those plug-ins can have new libraries and new compiler optimizations.”
Moreover, businesses can use Codon for the two prototyping and developing their apps. “A pattern we see is that men and women do their prototyping and testing with Python since it’s easy to use, but when press comes to shove, they have to rewrite [their app] or get anyone else to rewrite it in C or C++ to check it on a bigger details established,” says Shajii. “With Codon, you can stay with Python and get the greatest of both worlds.”
In terms of what’s upcoming for Codon, Shajii and his group are at the moment functioning on native implementations of broadly made use of Python libraries, as very well as library-unique optimizations to get a lot superior functionality out of these libraries. They also approach to develop a extensively requested attribute: a WebAssembly again finish for Codon to allow managing code on a Internet browser.
